To find novel factors required for gene regulation in the Drosophila embryo, we have analyzed mutants isolated in a screen for maternal factors required for embryo patterning performed in the Nüsslein-Volhard laboratory in Tübingen (Luschnig et al. 2004). In my laboratory, we studied 15 mutants that cause segmentation defects and examined gene expression patterns in mutant embryos. We selected two mutants that produce specific gene expression phenotypes. These genes have been mapped, isolated and characterized molecularly. One mutant disrupts the brakeless gene. Brakeless is a nuclear protein of unknown function. In brakeless mutant embryos, we observed expanded expression domains of the gap genes Krüppel (Kr) and knirps (kni). We found that Tailless-mediated repression of kni expression is impaired in brakeless mutants. Tailless and Brakeless bind each other in vitro and interact genetically.
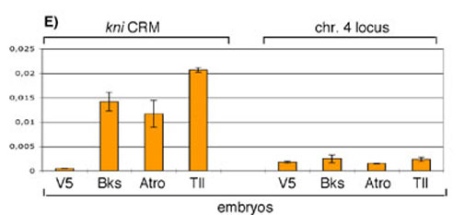
Brakeless is recruited to the Kr and kni enhancers (Fig 1), and represses transcription when tethered to DNA. This suggests that Brakeless is a novel co-repressor. Tailless and some other nuclear receptors also interact with the co-repressor Atrophin, the homolog of Atrophin-1 that causes the human neurodegenerative disease dentatorubral-pallidoluysian atrophy (DRPLA). We could show that Brakeless and Atrophin interact in vitro, and propose that they act together as a co-repressor complex in many developmental contexts. (Haecker et al. 2007).
The other mutant corresponds to a UDP-glucose:dolichyl-phosphate glucosyltransferase involved in N-linked protein glycosylation. In these mutants, gene expression in the posterior of embryos is affected due to reduced expression of the Caudal activator protein. The molecular mechanism behind this phenotype is under investigation.
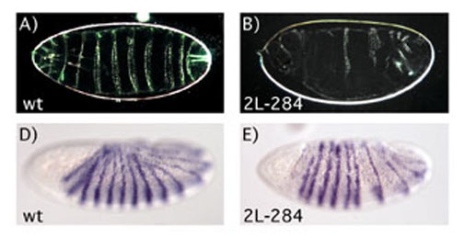
These studies have identified novel proteins that control gene expression, and will further increase our understanding of the developmental biology of the Drosophila embryo. We believe that additional coregulators can be identified from this collection of mutants, which may shed light on the mechanisms of gene and chromatin regulation that control development.
References
Haecker A., Bergman M., Neupert C., Moussian B., Luschnig S., Aebi M., and M. Mannervik. Wollknäuel is required for embryo patterning and encodes the Drosophila ALG5 UDP-glucose:dolichyl-phosphate glucosyltransferase. Development, 2008 135, 1745-1749.
Haecker A., Dai Q., Lilja T., Moussian B., Andrioli LP., Luschnig S., and M. Mannervik (2007). Drosophila Brakeless Interacts with Atrophin and is Required for Tailless-Mediated Transcriptional Repression in Early Embryos. PLoS Biol. 2007 Jun;5(6):e145.
Luschnig, S., et al., An F1 genetic screen for maternal-effect mutations affecting embryonic pattern formation in Drosophila melanogaster. Genetics. 2004. 167: 325-42.




