Role of chromatin in silencing and DSB-repair
The genomic DNA in eukaryotic cells is organized into a structure called chromatin. The basic unit of chromatin is the nucleosome in which 146bp of DNA is wrapped around a histone octamer. Posttranslational modifications of histones affect chromatin structure, which in turn affect processes like transcription and DNA-repair. We want to investigate how transcriptional silencing and DSB-repair is affected by chromatin structure.
We are studying silencing of the cryptic mating type loci (HM-loci) in K. lactis as a model for chromatin effects on transcription. Proteins binding to DNA sequences close to the HM-loci influence the establishment and maintenance of silencing. These so-called silencer binding proteins are different from the corresponding S. cerevisiae proteins and we are investigating how these proteins participate in silencing. In addition we are using strains with mutations in histones, specifically affecting positions subjected to posttranslational modifications, to elucidate the role of histone modifications in silencing and DSB-repair.
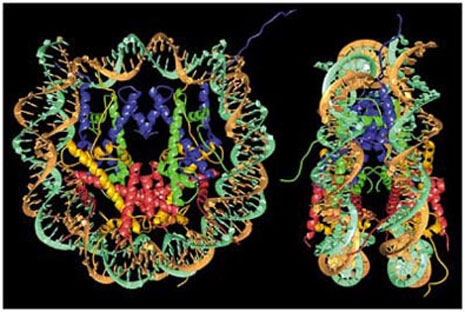
Front view (left) and side view (right) of 146bp of DNA wrapped around a histone octamer. Histone H2A (yellow), H2B (red), H3 (blue), H4 (green). Luger K. et al. 1997.
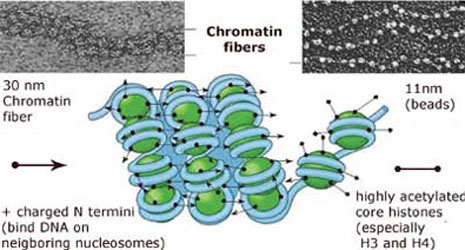
The DNA (blue) is wrapped around a histone octamer (green), which is organized into higher order structures. The picture is adapted from http://sgi.bls.umkc.edu/
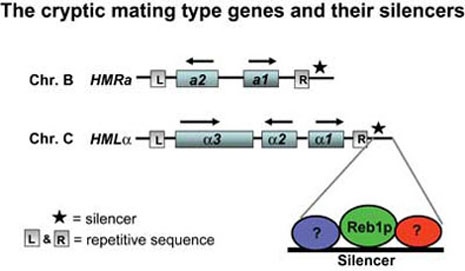
These loci encode genes specific for the two mating types (a and alpha), but are not expressed due to transcriptional silencing. The silencers are denoted with an asterisk. The HML silencer is enlarged showing protein binding sites.
L and R= repetitive sequence used for resolving recombination intermediates during mating type switch.




