Research group Arne Elofsson's research grop
Combining large scale life-science data with artificial intelligence is crucial for the continued progression of our understanding of the molecular processes that govern life. We propose developing novel deep-learning methods to provide an unprecedented accurate description of the human proteome.

Group description
Predicting protein-protein interactions using AI
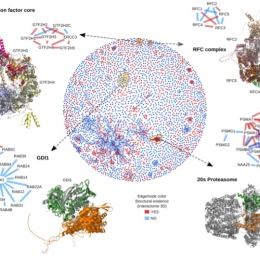
Central to the methods we will develop is that they can efficiently utilise both annotated and unannotated biological data using self-supervision. We bring together four groups with complementary skills with the ultimate goal of providing an unprecedented detailed description of the human proteomic landscape. Starting from a novel set of long-read transcripts combined with existing large-scale proteomic analysis, we will first use novel machine learning methods to identify proteoforms, i.e. splice variants and post-translational modifications. These proteoforms will then be the basis for the identification of protein-protein interactions. Two approaches will be developed; first, language models will identify permanent and transient protein-protein interactions from the proteoforms. Secondly, we will use structural modelling by Alphafold to provide atomistic models of interacting protein pairs. Finally, the project will provide atomistic models of these proteoforms and their interaction partners, including large complexes. We will validate these complexes using native mass spectrometry. We believe that this project is extremely timely, given that AlphaFold was the scientific breakthrough in Science 2021 and the method of the year in Nature Methods, and that KAW has invested heavily in data-driven life science (DDLS) and compute power (Berzelius computer at NSC)
We are applying this philosophy primarily to the studies of two very important classes of proteins: transmembrane proteins and repeat domain containing proteins. Both classes of proteins are important drug-targets and/or central to diseases. Our studies use a broad range of techniques. We are primarily using bioinformatics and other computational methods, but also to an increasing fraction biochemical and other experimental techniques.
Group members
Group managers
Arne Elofsson
Professor of Bioinformatics

Members
Matteo Tadiello
Doktorand

Samuel Fromm
Postdoc

Petras Kundrotas
Researcher

Oxana Lundström
PhD student

Sarah Narrowe Danielsson
PhD student