Research project Predictions of protein-protein interactions by PconsDock, a fold-and-dock algorithm.
Today it is possible to predict the structure of most proteins accurately, but proteins do not act alone. Prediction of protein interactions has been a more difficult challenge than predicting the structure of individual proteins, and progress has been limited.
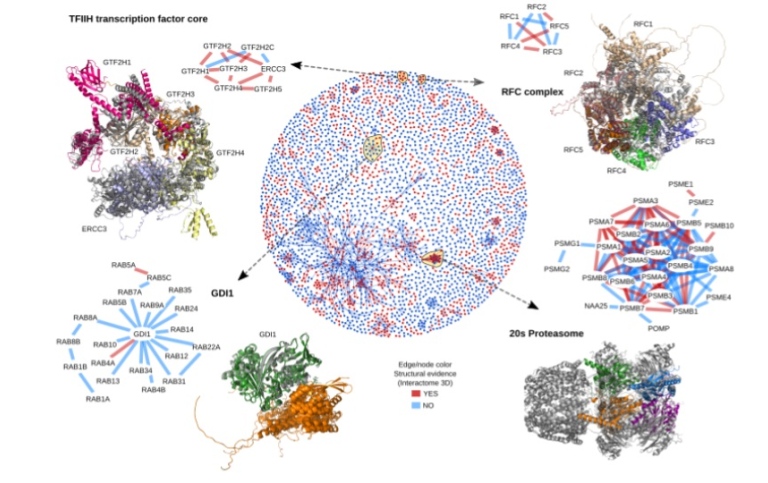
Many different techniques have been developed, but none of these is very accurate. In the last decade, de novo protein structure prediction accuracy for individual proteins, using co-evolution and deep learning, harvesting the information from large multiple sequence alignments. These methods can, in principle, also be used to extract information about protein-protein interactions. However, so far, success has been limited to a handful of cases. Here, we propose to utilise the latest progress for predicting individual protein to improve protein-protein docking using PconsDock. Our novel PconsDock protocol can accurately predict the structure of protein pairs where alternative methods fail. However, still there exists a large set of protein-protein pairs where PconsDock fails. Therefore, we aim to continue developing PconsDock by (i) investigating improved methods to identify interaction protein sequences and (ii) developing improved deep learning methods to identify the contacts accurately. We believe that the predicted structures will contribute significantly to the set of tools available for understanding cellular and evolutionary processes, providing an essential step towards a structural map of an entire cell.
Project members
Project managers
Arne Elofsson
Professor of Bioinformatics


