Research project Transcription, coregulators and histone acetylation
Gene transcription is essential for cell identity and its misregulation causes disease. This project explores the connection between transcription and chromatin modifications that make genes accessible to the transcriptional machinery.
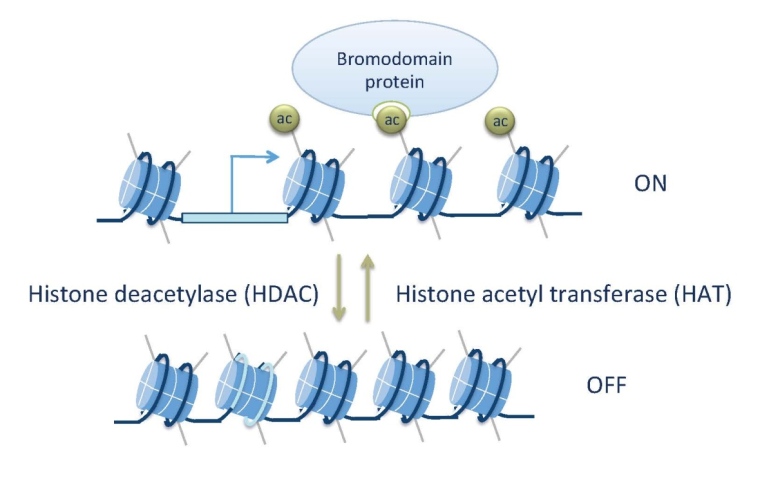
In this project we characterize the role of transcriptional coregulators and histone acetylation in gene regulation. Coregulators are proteins that do not bind to DNA, but that facilitate communication between sequence-specific transcription factors and the RNA polymerase machinery (Mannervik et al. 1999). One function of coregulators is to modify chromatin by acetylating or deacetylating histones. A number of histone acetyltransferases (HATs) and deacetylases (HDACs) are conserved between organisms as diverse as yeast and human. We are using the Drosophila embryo as our “test tube” to characterizee them and histone modifications in vivo,. Using genomic technologies, genetic tools and transgenic embryos, we can obtain an understanding of the molecular functions of these proteins in development. Given that chromatin regulators are deregulated in many cancers, these studies will also contribute to an understanding of how they can cause uncontrolled cell proliferation and cancer.
Project description
Is histone acetylation necessary for transcription?
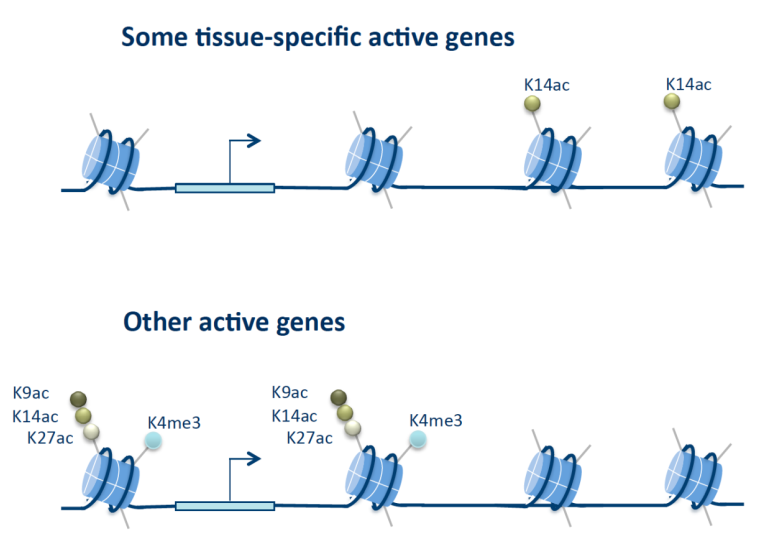
To directly probe the role of histone modifications in transcription we have used a histone replacement strategy in Drosophila where the endogenous histones are replaced by transgenic histones. Individual amino acids in the transgenic histones can be altered to study the consequences for organismal development and fitness. We have replaced histone 3 lysine 14 (H3K14) for arginine (H3K14R), since this lysine is acetylated but not methylated (although low levels can be detected in mammals). We discovered that H3K14 is required for expression of some tissue-specific genes that contain H3K14ac, but lack other histone acetylations and methylations associated with most active genes. Absence of H3K14ac and defective gene expression causes lethality and defective wing patterning. We found that the SWI/SNF protein Brahma (Brm) recognizes H3K14ac, that brm acts in the same genetic pathway as H3K14R, and that chromatin accessibility at H3K14ac-unique genes is decreased in H3K14R mutants. These results show that acetylation of a single lysine is essential at genes devoid of canonical histone marks and uncover an important requirement for H3K14 in tissue-specific gene regulation (Regadas et al. 2021).
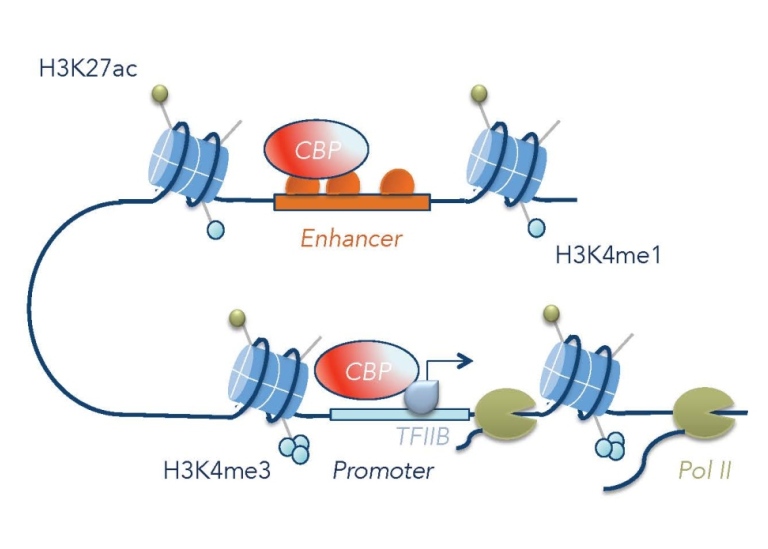
The p300/CBP coactivator
The p300 and CBP co-activators are histone acetyltransferases and central regulators of transcription in metazoans. They preferentially acetylate histone 3 lysine 27 (H3K27), H3K18 and H4K8. The Drosophila homolog of p300 and CBP is known as Nejire, or CBP. We have shown that Nejire/CBP controls signaling by the TGF-ß molecule Dpp in the Drosophila embryo by multiple mechanisms (Lilja et al. 2003, 2007). The genomic occupancy of p300/CBP detected by ChIP-seq experiments can be used to identify transcriptional enhancers (e.g. Negre et al. 2011). However, our studies in Drosophila embryos suggest that there is a preference for some transcription factors in directing CBP to the genome (Holmqvist et al. 2012). ChIP-seq studies of CBP in Drosophila S2 cells showed that CBP occupies many genomic elements, not only enhancers (Philip et al. 2015). We found that CBP has two important functions at promoters of active genes, where it helps in recruiting RNA polymerase II by interacting with the general transcription factor TFIIB, and in releasing promoter-proximal paused RNA polymerase into productive elongation through acetylation of the +1 nucleosome (Boija et al. 2017). Recruiting the CBP HAT domain to a silent gene promoter or enhancer through a fusion with catalytically dead Cas9 can be sufficient for gene activation, but not at all silent genes (Sajwan and Mannervik 2019). Although p300/CBP occupancy in general correlates with gene activation, it can also be found at silent genomic regions, which does not result in histone acetylation (Holmqvist and Mannervik 2013). Polycomb-mediated H3K27me3 is associated with repression, but does not preclude p300/CBP binding. We have recently discovered that CBP has a non-enzymatic function at Polycomb-repressed genes that is required for Polycomb-mediated gene silencing (Hunt et al. in prep.).
HDACs in transcription
What is the direct role of histone acetylation in transcription? To answer this question we have treated cells for a short time period (10 min) with an HDAC inhibitor and studied the direct effects on transcription by precision run-on sequencing (PRO-seq). We found that increased acetylation after HDAC inhibition results in release of promoter-proximal paused Pol II into productive elongation at genes that are already highly acetylated before treatment (Vaid et al. 2020). This shows that histone acetylation faclitates Pol II transcription beyond the +1 nucleosome.
An Ebi-HDAC3 corepressor complex
The Snail repressor protein is required for mesoderm formation through its downregulation of neuroectoderm-specific genes. We found that in Drosophila embryos lacking the maternal contribution of the Ebi protein, Snail target genes are de-repressed leading to mis-expression of neuroectoderm specific genes in the presumptive mesoderm. The Ebi protein is the homolog of mammalian TBL1 that associates with HDAC3 and the co-repressors SMRT and NcoR. We have identified a motif conserved in all insect Snail-related proteins. This Ebi interaction region constitutes a potent repression domain. Ectopic expression of full-length Snail and a mutant Snail lacking the Ebi interaction domain in transgenic embryos demonstrated that the Ebi interaction domain is essential for Snail function in vivo. Together, these results show that Ebi is a corepressor required for Snail activity (Qi et al. 2008).
Although Ebi associates with HDAC3, the role of histone deacetylation in Snail-mediated repression is not known. We have therefore targeted HDAC3 by homologous recombination and generated point mutants that abolish catalytic activity. Interestingly, we found that HDAC3 can both activate and repress transcription and that these functions can be genetically separated. HDAC3 catalytic activity is not necessary for Snail-mediated repression, but is required for gene activation by HDAC3 (Min et al, manuscript in prep.)
The Brakeless and Atrophin co-repressors and neurodegeneration
To find novel factors required for gene regulation in the Drosophila embryo, we have analyzed mutants isolated in a screen for maternal factors required for embryo patterning performed in the Nüsslein-Volhard laboratory in Tübingen (Luschnig et al. 2004). In my laboratory, we studied 15 mutants that cause segmentation defects and examined gene expression patterns in mutant embryos. One mutant disrupts the brakeless gene. Brakeless is a nuclear protein of previously unknown function. We found that Brakeless is a novel co-repressor required for function of the Tailless repressor.
Tailless and some other nuclear receptors also interact with the co-repressor Atrophin, the homolog of Atrophin-1 that causes the human neurodegenerative disease dentatorubral-pallidoluysian atrophy (DRPLA). We could show that Brakeless and Atrophin interact in vitro, and propose that they act together as a co-repressor complex in many developmental contexts. (Haecker et al. 2007). We found that Brakeless can activate some genes and repress others, suggesting that it may switch between a co-activator and co-repressor function (Crona et al. 2015). Atrophin modulates the activity of Drosophila GAGA factor (Trithorax-like, Trl), and functions in developmental gene transcription (Yeung et al. 2017).

